Portfolio

Description of select projects
- Establishing a CRISPR-based
variant-to-function assay, applied to CD19 enhancer
Learn more - Full-stack web development of
transcription-factor motif database
Learn more - Applying variant-to-function
assay to identify therapeutic targets for beta-hemoglobinopathies
Learn more
CRISPR-Millipede
Establishing a CRISPR-based variant-to-function assay to investigate a novel CD19 enhancer to inform CAR-T therapies
This is the first chapter of my PhD thesis work, where I established a high-resolution
CRISPR-based variant-to-function framework to understand non-coding regulatory sequences.
This project led to a first-author manuscript (provisionally accepted at
Nature
Communications) and is available as a preprint on bioRxiv.
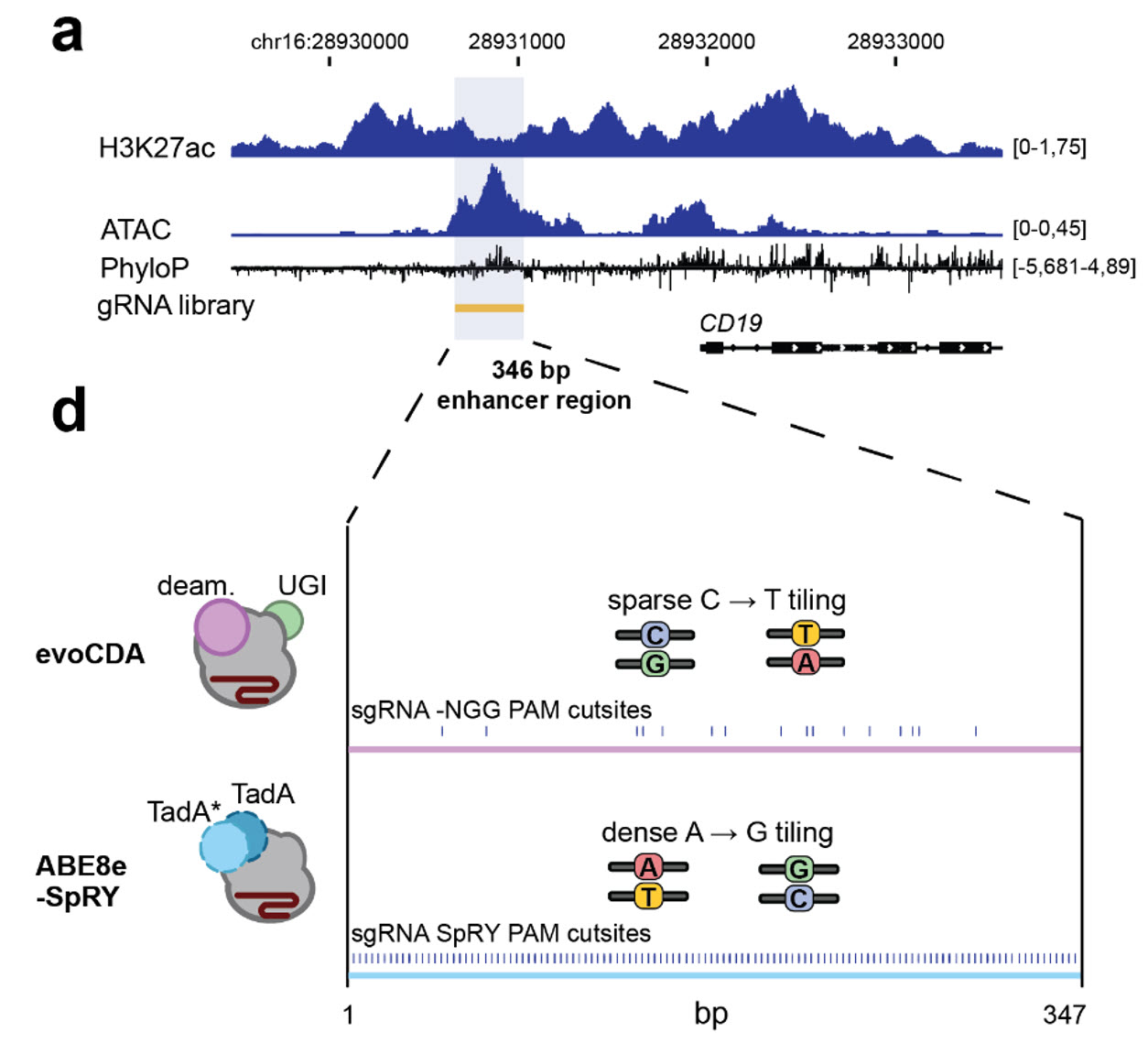
We investigated a putative regulatory element upstream of the CD19 gene, a B-cell marker and immunotherapy target in pediatric and adult B-cell acute lymphoblastic leukemia (B-ALL) and lymphoma. The candidate regulatory element was identified based on the presence of a highly evolutionary conserved and open chromatin region in proximity to the CD19 gene, as indicated by ATAC-seq and H3K27ac ChIP-seq.
I have designed an experimental approach expanding on traditional base-editing
mutagenesis screens. Dense base-editing mutagenesis allows single-variant
mutations to
be installed throughout a region of interest, which allows the assessment of each
mutation's phenotypic impact based on fluorescence-activated cell sorting (FACS) of CD19
expression.
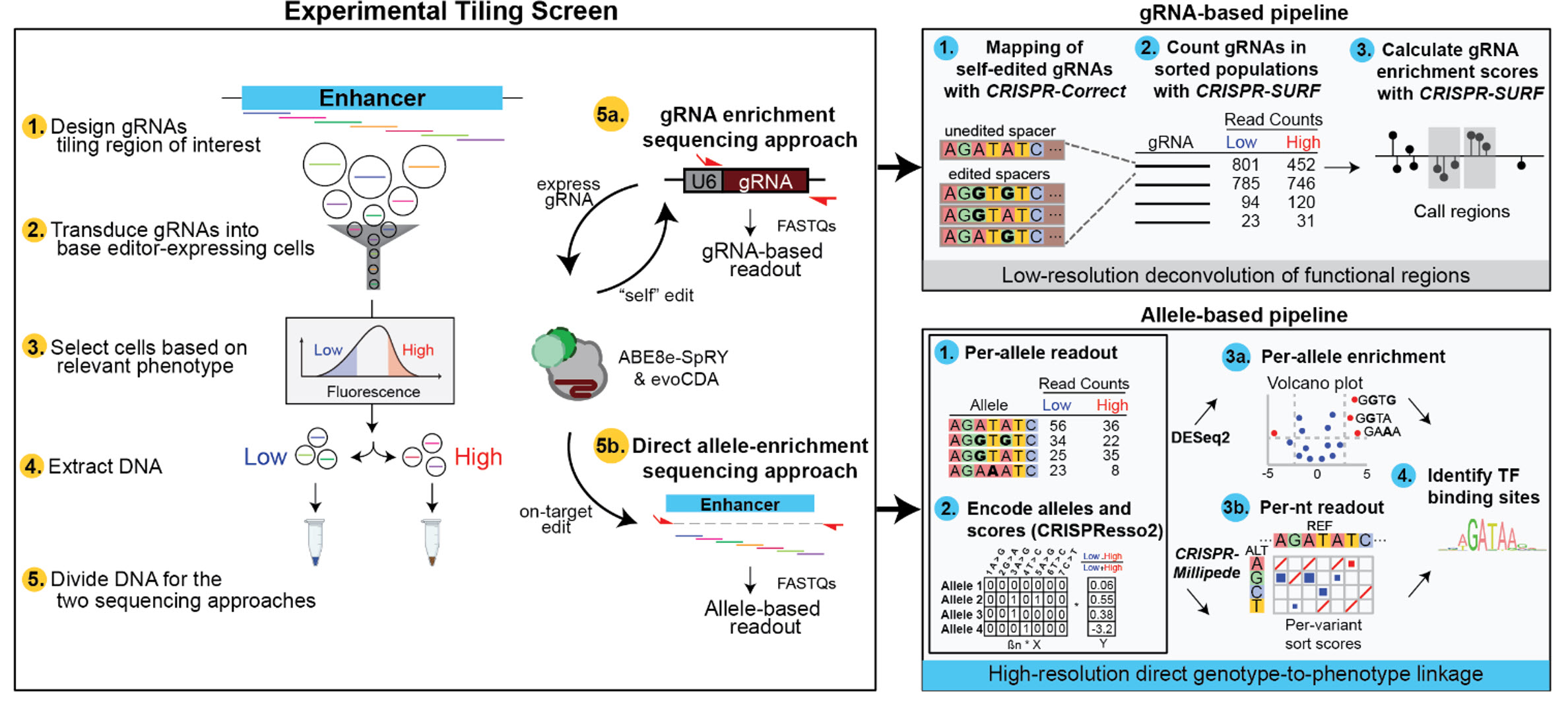
After sorting, genomic DNA is split between two library preparation approaches, the
standard approach that performs amplicon-sequencing of the CRISPR guide RNA integrant,
and the second approach which performs direct amplicon-sequencing of the edited genomic
locus. Measuring the count of either guide RNA (from the former approach) or installed
target allele (from the latter approach) in each sorted cell population can be used to
measure phenotypic enrichment of guide RNAs or alleles.
We go a step further to generate high-resolution per-nucleotide scores.
We perform
Bayesian regression modelling of the target allele enrichment scores using the
millipede tool to generate per-nucleotide enrichment scores.
SOFTWARE PRODUCED:
For this framework, I have developed two primary computational tools. First, I developed
CRISPR-Correct, which is a CRISPR guide RNA mapping Python tool that handles
self-editing of
the protospacer via a hamming-distance approach https://github.com/pinellolab/CRISPR-Correct.
This framework is also available to run at scale on Cromwell via Broad Institute's Terra
platform: https://github.com/pinellolab/CRISPR-millipede-pipelines.
Second, I developed CRISPR-Millipede, which is a Python tool that performs Bayesian
regression modelling of allele enrichment scores to generate per-nucleotide enrichment
scores https://github.com/pinellolab/CRISPR-millipede-target.
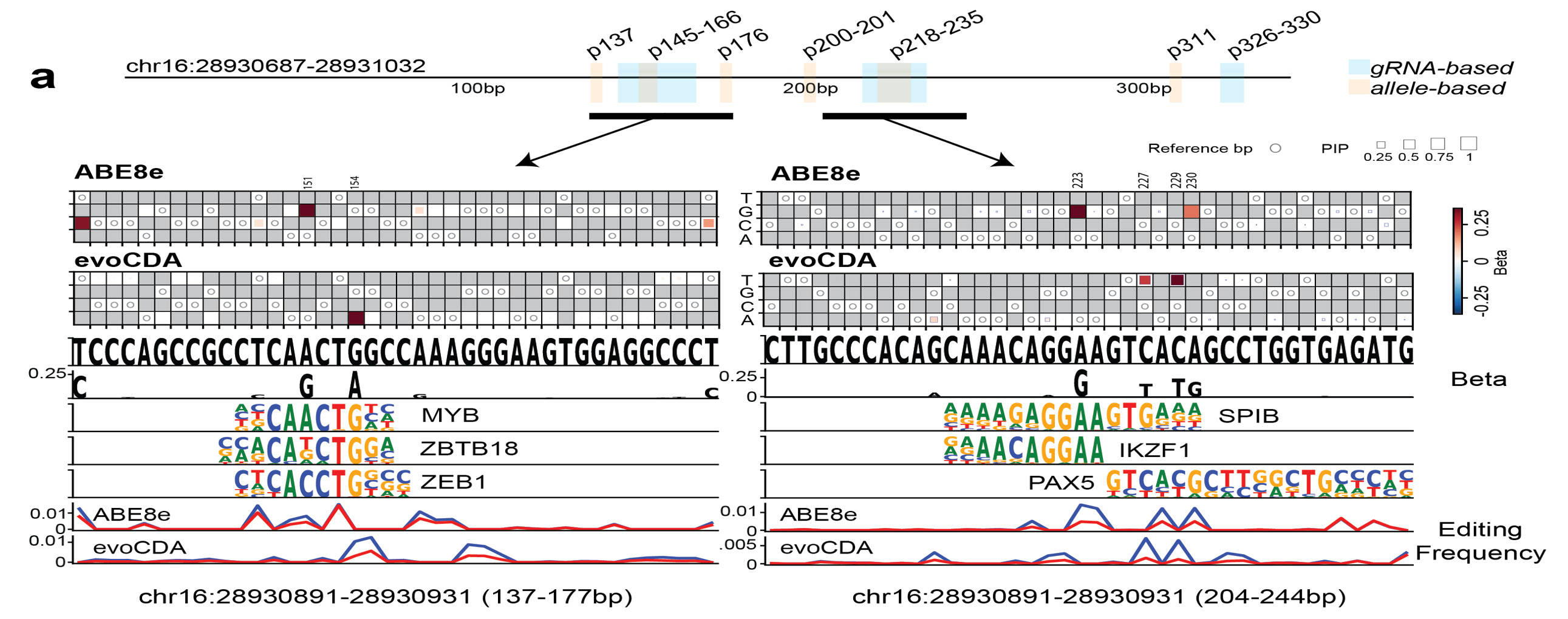
The higher-resolution single-nucleotide scores can be used to help disambiguate
between
annotated
transcription-factor binding motifs, thereby supporting the identification
of
novel
binding
sites in non-coding sequences. For instance, we have identified both a MYB site
and an
IKZF1::PAX5 binding site motif, which are known B-cell regulators.
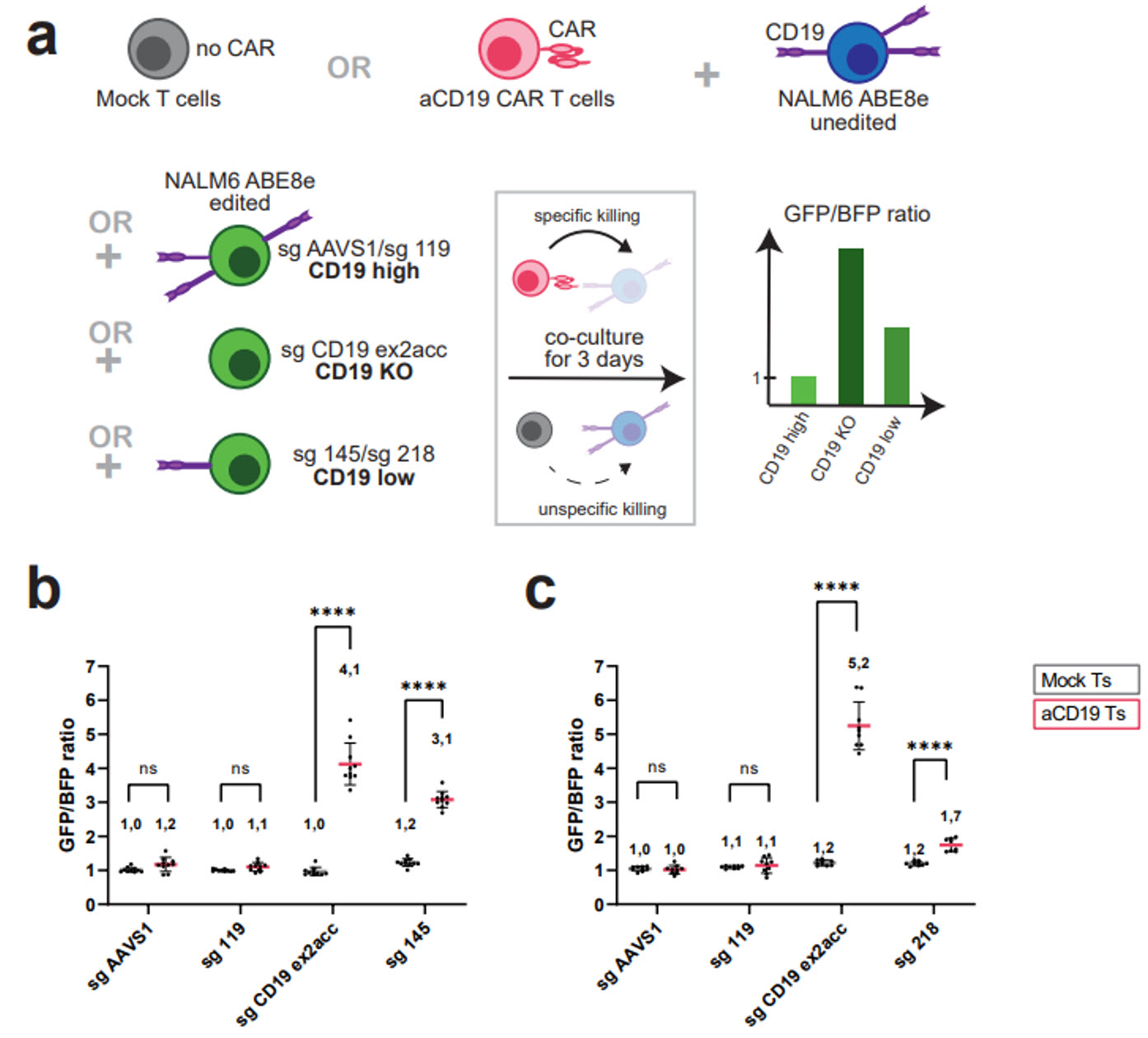
We were interested in if these variants may effect CAR-T therapies.
CD19 is a target of
CAR-T cell therapy in B-cell malignancies. Despite initial high
success rates, ~35% of B-ALL cases treated with aCD19 CAR-T relapse, with a subset
due
to loss or downregulation of the CD19 antigen. In a CAR-T targeting assay, we show
that
functional variants in the CD19 enhancer cause increased resistance to CAR-T
targeting.
MotifCentral.org
Developing a full-stack web-application to visualize transcription-factor binding motifs.
MotifCentral.org is a full-stack web
application to display a database of transcription-factor
binding motifs fit by the ProBound statistical model on SELEX-seq datasets. The
architecture
is comprehensive: accessing stored motif information on PostgreSQL
(hosted
on Amazon RDS)
via Spring-Boot (Java) RESTful web-services (hosted on Amazon
Elastic Beanstalk) and finally
displayed on an Angular 7 web page (hosted on Amazon
CloudFront, with the networking handled
by Amazon VPC and Amazon Route53). The curation and annotations of the
model are also done
via an administrative privilege on the web application – administrators can log in and
authenticate themselves with Amazon Cognito. Users are synced between the Amazon
Cognito
storage and the PostgreSQL database via an Amazon Lambda trigger upon
user
sign-up. All code
was developed before the release of ChatGPT.
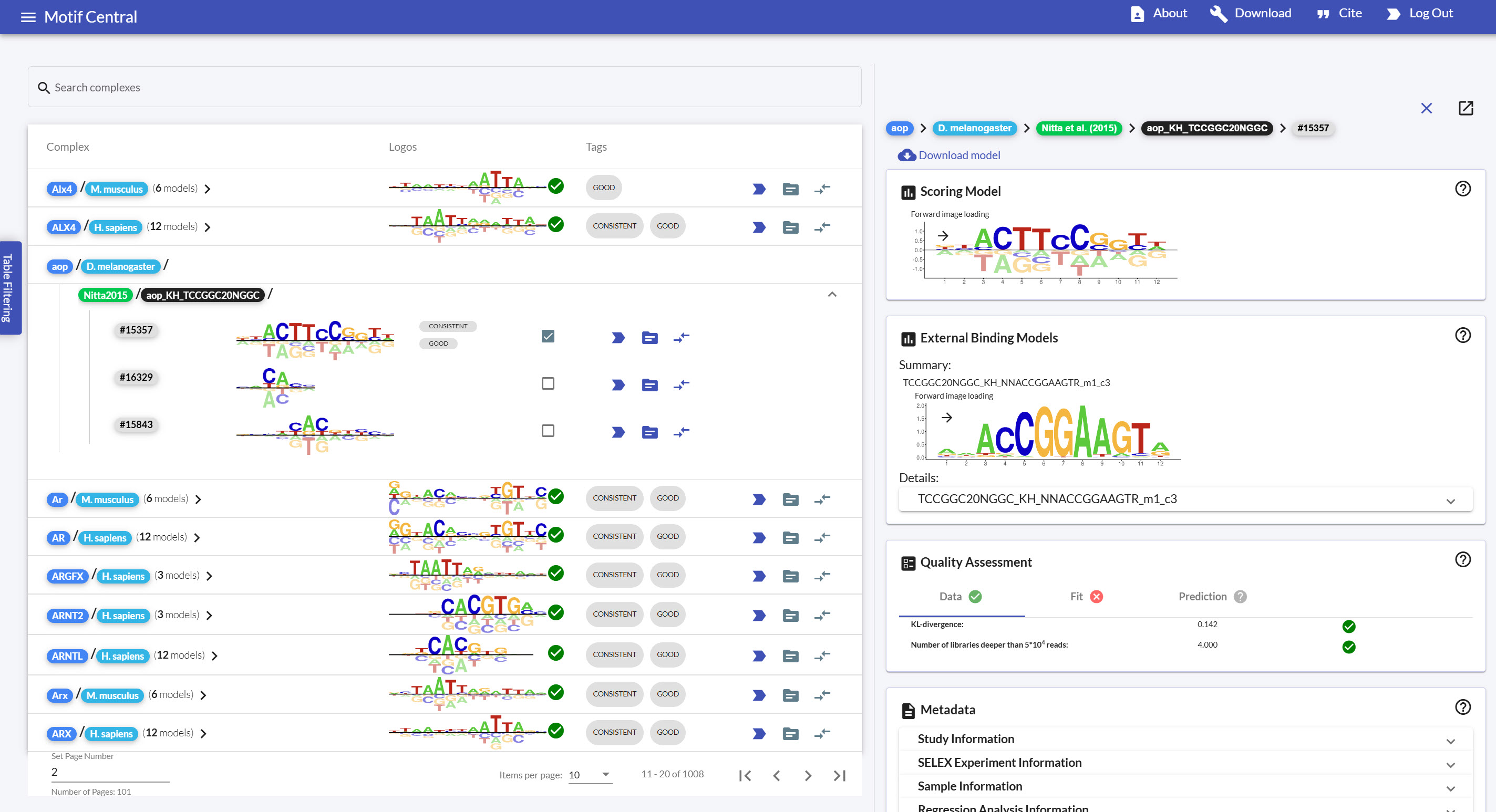
Screenshot of MotifCentral.
The Front-End (with Angular 7)
To describe the implementation of the front-end Angular web-app, it is essential to understand the general idea of Angular – specifically, Angular web-apps are organized into components that represents different “sections” of the web page. Components could be reusable across different areas of the site or only have a single instance in the site. Components could be hierarchical (i.e. a component may have several sub-components), and components can communicate and pass data to each other in multitudes of ways. For example, here is an an illustration of some components from the web-page:
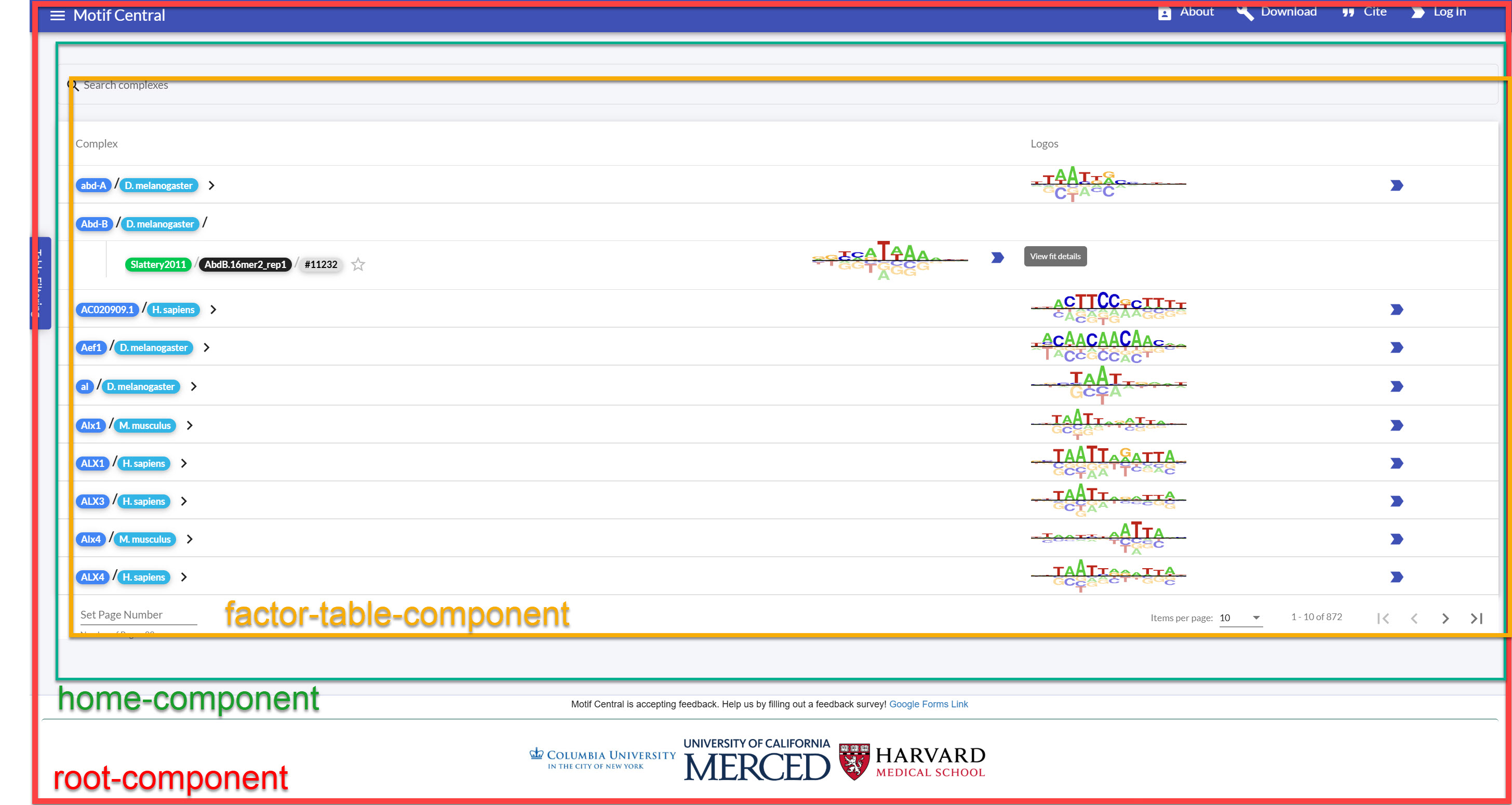
Illustration of Angular components in MotifCentral.
You have the “root-component” which contains the logic for the navigation bar, the
footer
bar, then the “content” of the page that depends on the URL (known as the
router-outlet), see the HTML portion of the root-component here:
https://gist.github.com/CodingBash/e615ab8c8c88a5b1168477ef1fe9e943.
For example, the component that the router-outlet generates at the root URL (i.e.
motifcentral.org) will display the HomeComponent according to the routing specification
loaded at app-routing.module.ts:
https://gist.github.com/CodingBash/548d025a9029dd4aad7a33e53c07eeb7.
The home-component primarily loads the factor-table component and helps communicate data
between the factor-table-filter component for table filtering and between the
in-page fit-details page to display the motif fit for a particular transcription factor
if
selected. This page contains some extra logic for displaying items necessary for the
administrator view only if the user identified is an administrator:
https://gist.github.com/CodingBash/f91d87ca39c67e08de57382e47f92d40
Of course, the first step is to retrieve all the relevant information: the list of all
transcription factor models as well as the user information. When the home-component is
loaded, one of the first functions called is the ngOnInit function
(https://gist.github.com/CodingBash/f91d87ca39c67e08de57382e47f92d40#file-public-view-component-ts-L51).

HomeComponent ngOnInit function - retrieval of user and fit information
The first step is to retrieve the user information via the UserPrincipalService. Services in angular are general objects used to retrieve, send, and/or handle data that can be “dependency injected” into components for usage. The UserPrincipalService simply contains a singleton object to store the user state as a BehaviorSubject (https://github.com/CodingBash/cellx-angular-front-end/blob/master/src/app/services/user-principal.service.ts#L11) which is a type of object allowing asynchronous access of the user state in an event-driven manner (i.e. if the user state changes, all “listeners” of the BehaviorSubject object will be triggered). The user state is set in the component of the “login” page, which uses the authentication token returned from Cognito to further access the user information (https://gist.github.com/CodingBash/459458ef3b477af162539ac896e056c3#file-login-component-ts). The token is then sent with every HTTP call to back-end web-service to be validated by REST service endpoints requiring authorization (discussed in back-end section). If an attempt to call an authorized endpoint is made with a non-admin user token, expired token, or fabricated token, the REST endpoint will return an “HTTP 403 Unauthorized” (this will be in the back-end description section).
Redirect to login screen from AWS Cognito. If login successful, Cognito redirects back to MotifCentral with a user token.
After retrieving the user information, the home-component then retrieves the
transcription-factor information
(https://gist.github.com/CodingBash/f91d87ca39c67e08de57382e47f92d40#file-public-view-component-ts-L65).
which uses the factors-service
(https://gist.github.com/CodingBash/f91d87ca39c67e08de57382e47f92d40#file-factors-service-ts)
to make REST calls to the appropriate endpoints depending on the environment
(development,
test, production).
The next important logic in the home-component page is the table filtering
(https://gist.github.com/CodingBash/f91d87ca39c67e08de57382e47f92d40#file-public-view-component-html-L24).
The filter was surprisingly more challenging to implement, as the selection of the
species
or the study must be communicated to the factor-table (through the home-component) to
filter
the motif fits with the selected attributes. Additionally, information from the
factor-table
must be communicated back to the filter component to display the available
species/studies
to select and the total number of motif fits available. This is the advantage of having
a
parent component to handle data communication (in this case, the home-component) using
@Input and @Output directives (https://v17.angular.io/guide/inputs-outputs). The
filter-component
needed to be handled in the filter-component to handle when to trigger a change for the
components to be updated
(https://gist.github.com/CodingBash/fdb854a485e3186ec3d3ec7adcc799ce#file-factor-table-filter-component-ts-L87).
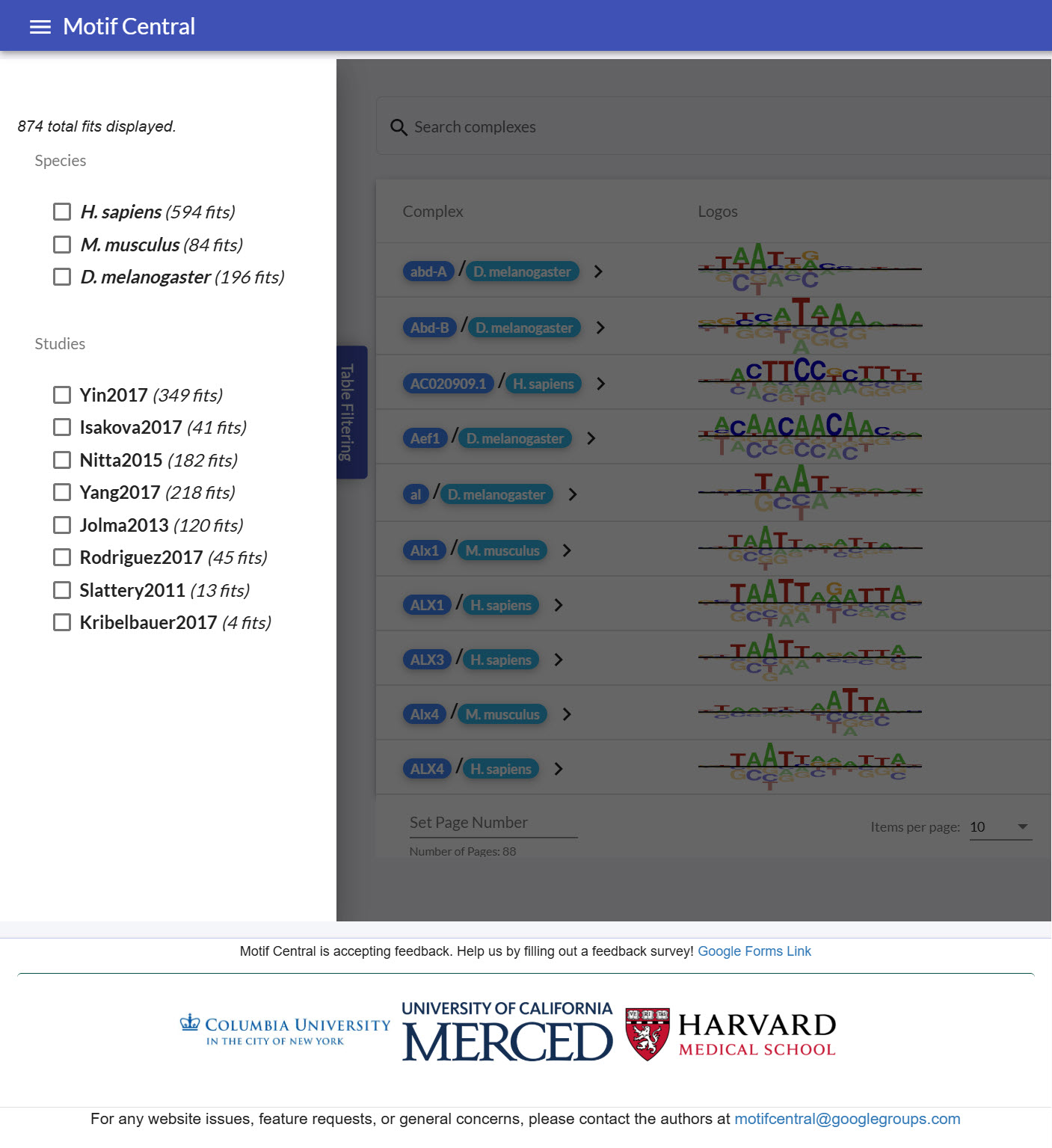
Display of filter component in MotifCentral
There are several more logic on displaying the table of factors that won’t be discussed in-depth. For example, displaying the motif logo image has some tricky optimizations that were performed. In the default implementation of the table, all images would be loaded at once, causing the website to become quite slow due to the simultaneous loading and the traffic to the file loading REST service. Therefore, images are only loaded if the element is seen in the screen viewpoint (https://gist.github.com/CodingBash/2da85d53b958de164b4b7c16dbe49b90).
Dynamic loading of images
The last primary aspect of the front-end application is displaying the details of a motif fit when a motif is selected from the home table. The page makes the most of components from the Angular Material UI library (https://material.angular.dev/components/categories) to display pop-up modals, accordion displays, cards, etc.
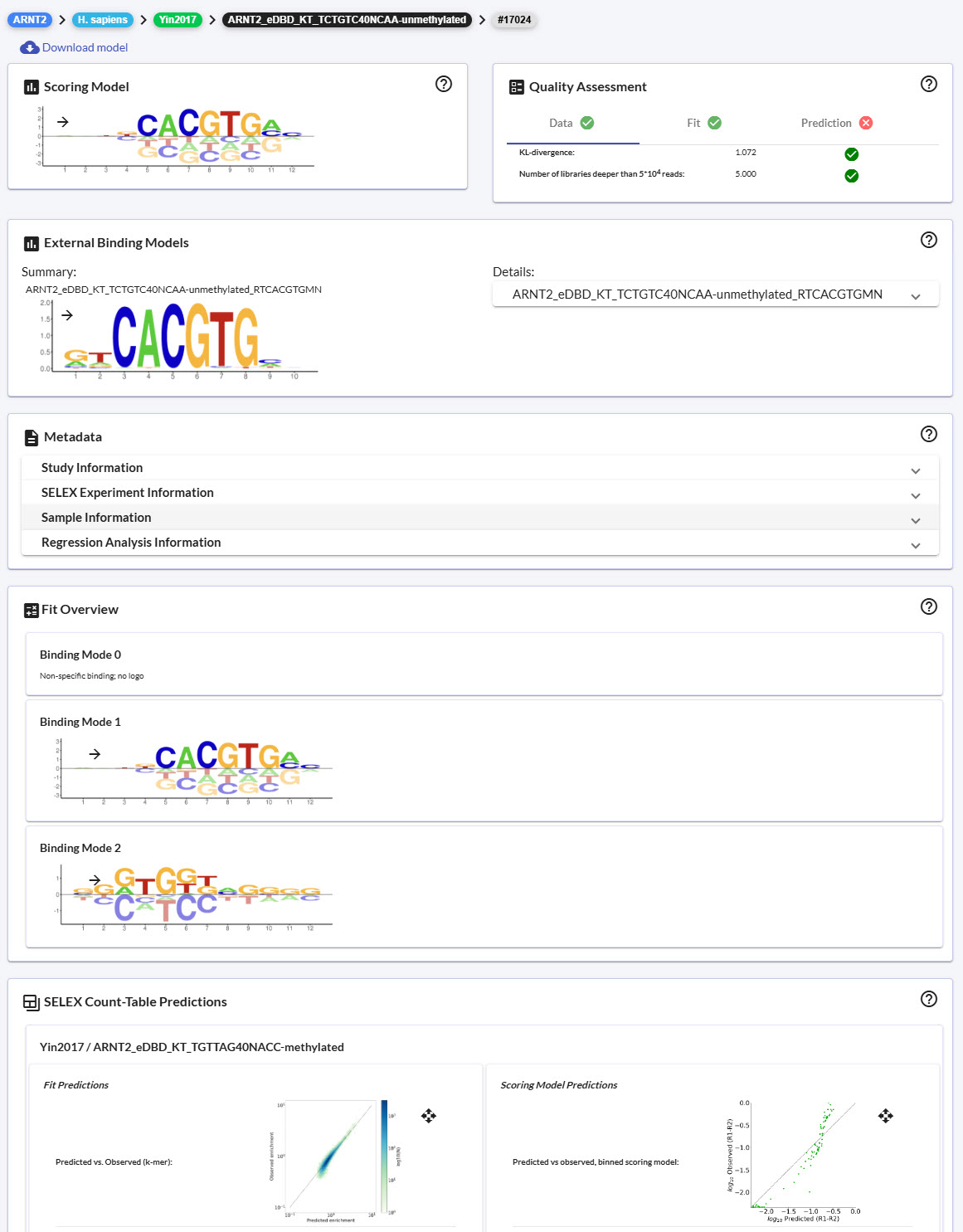
Fit details page
Each section of the details page has its own component (i.e. scoring model, quality assessment, etc.). The fit-details-component is the parent component that organizes each of these individual components on the page. These individual components are responsible for making the appropriate REST calls to retrieve data along with populating the Angular Materials component with the retrieved data.
List of all the individual components in the fit details page
It is seamless to deploy the Angular website to AWS using AWS CodePipeline’s continuous deployment service. The only manual step to deploy is to push the code to the prod-deployment branch on GitHub, which then triggers CodePipeline to retrieve the source code, build the Angular deployable (with the current environment specification so that all REST calls in the Angular app points to the production endpoints), and store the web files in an Amazon S3 bucket. Amazon CloudFront services the web files to the browser at the configured DNS specified as motifcentral.org, where the domain is handled and managed by Amazon Route53.
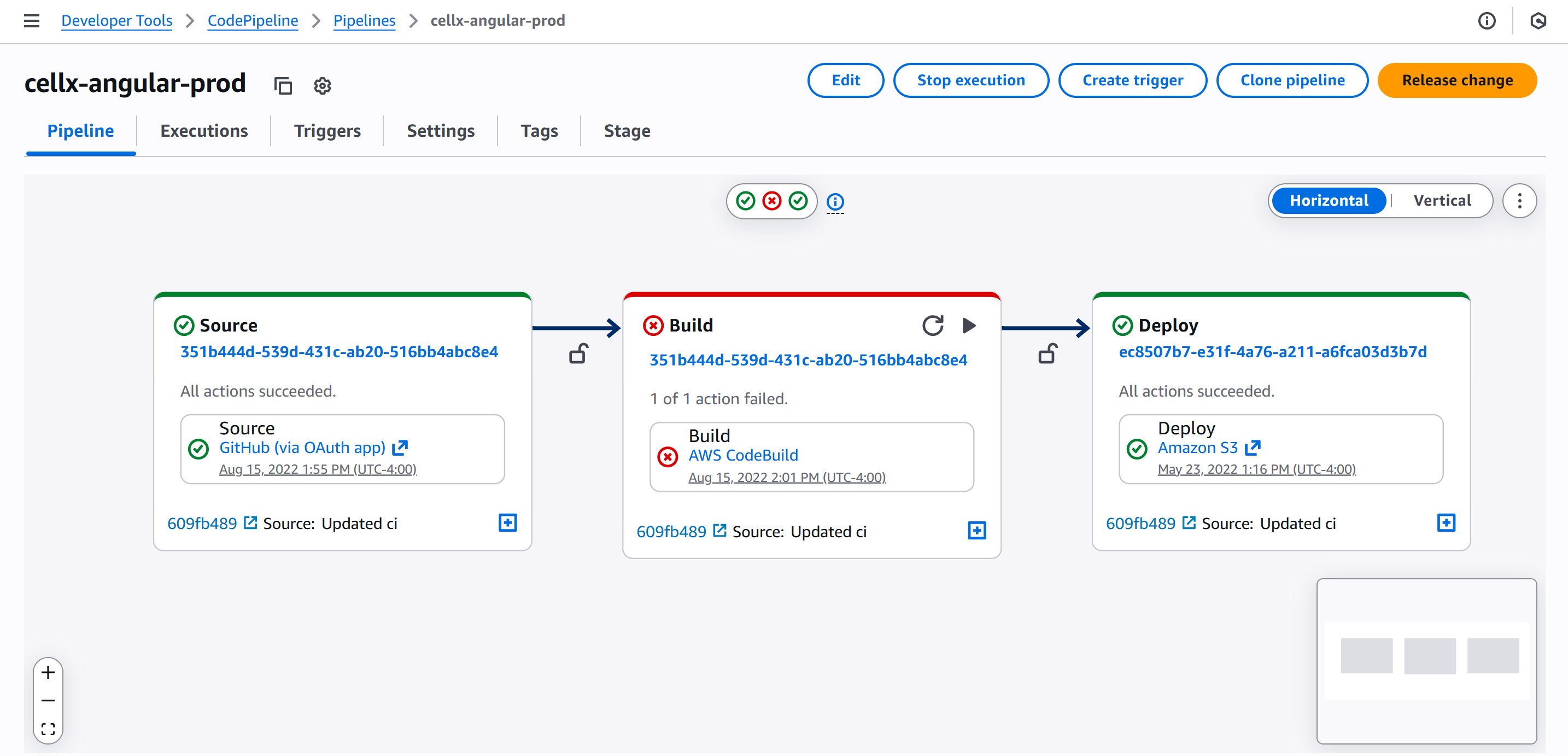
CodePipeline interface for deployment of the Angular web page to AWS S3 and CloudFront
The Back-End (with Spring Boot, AWS Lambda, and PostgreSQL with AWS RDS)
As mentioned previously, the Angular front-end services make several REST calls to
retrieve
data. The REST services are developed in Java using the Sprint-Boot framework, although
most
web-frameworks operate in a similar manner, though I find Spring-Boot to be most
comprehensive and readable due to its wide usage in large enterprises (i.e. Netflix,
AirBnB,
Uber,
PayPal, etc.).
Spring-Boot REST services are relatively simple in design. The classes that contain the
specifications for the endpoints are termed “Controllers”. For instance, I have a
controller
called “ComplexSpeciesTreeController” that contains endpoints to retrieve relevant
information about the transcription factors
(https://gist.github.com/CodingBash/8217c2019f917f630fd8de51d6ee5f22).
An endpoint is
defined as a function that is annotated with @RequestMapping which specifies the URL
path
(i.e. "/taxcomplexes/search”
https://gist.github.com/CodingBash/8217c2019f917f630fd8de51d6ee5f22#file-complexspeciestreecontroller-java-L75)
and the request method (i.e. HTTP GET). The function can also be annotated based on the
authority of the logged-in user using the @Secured annotation, which specifies rules to
allow access to the user. Configuration is required to perform user authentication,
specifically providing the endpoints to interact with AWS Cognito for user
authentication,
and storing user session in an in-memory database using Redis. Spring-Boot handles the
specific operations for interacting with Cognito and Redis.

Example of an endpoint definition in Spring-Boot
Another important software design pattern in Spring-Boot (and also in Angular and
several
other modern frameworks) is dependency injection. Similar to dependency injection of
“services” to retrieve data in Angular, Spring-Boot effectively has the same design of
accessing “services” to retrieve data. Specifically, controllers access services to
retrieve processed data, and services retrieve and process the raw data from
dependency
injected “repositories” classes that performs the SQL or REST transactions
(https://gist.github.com/CodingBash/2ac7e305c747ff220d44d0909dc4a71d).
I also include an endpoint that is used to stream the motif logo images from Amazon S3 to
the front-end
while
handle authentication (https://gist.github.com/CodingBash/35a40b34240293ea3c5db470a80a8b2c).
No credentials is required for the web-service to access S3 as they exist on the same
private VPC, therefore the S3 URLs cannot be accessed from the internet. If a service
performs multiple repository calls to access SQL data, Spring-Boot can aggregate all
calls
as an SQL transaction to ensure data reliability.
These repositories could be accessed across several services, and services could be
accessed
across several controller endpoints, therefore dependency injection typically allows a
single
instance of the class to be shared. I find Spring Boot applications to be much more
readable
and reliable compared to event-driven and dynamic typecasting frameworks such as
Node.JS,
which may be why Spring-Boot is preferred in many enterprises.

Example of a repository function performing an SQL command
Deploying REST services are also done using CodePipeline, where a commit to the production GitHub branch triggers a build then deployed onto Elastic Beanstalk. Of course, each step requires configuration to specify the build commands and the deployment operations, though once the configurations are complete, deployment is more quick and reduces chances of mistakes during deployment due to automation.
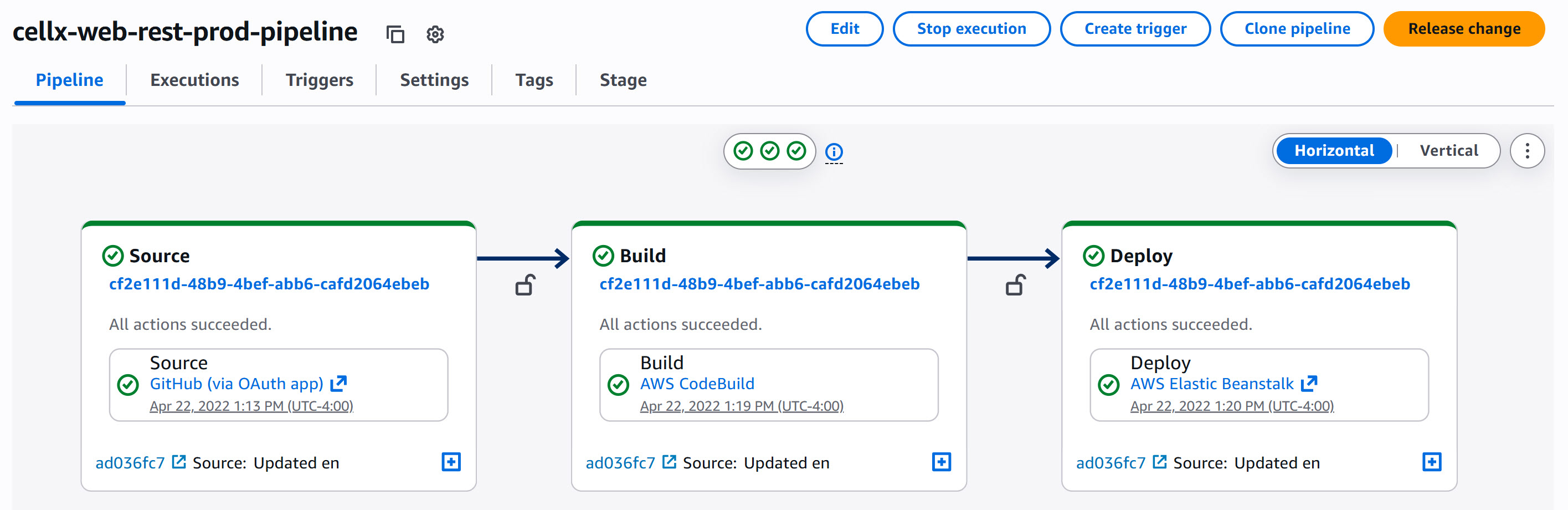
CodePipeline interface showing deployment of REST service to AWS Elastic Beanstalk
Therapeutic genome editing for sickle-cell disease
Multi-modal dissection of sequences regulating fetal hemoglobin for optimal therapeutic genome editing.
In this project, I established a multi-modal variant-to-function framework to identify therapeutic targets for beta-hemoglobinopathies in Dr. Luca Pinello and Dr. Dan Bauer labs, specifically expanding upon the first CRISPR gene therapy for sickle-cell disease (CASGEVY) discovered by Dr. Dan Bauer.
My PhD Trajectory
I want to give some background on the significance of this project in terms of scientific
training. During my rotations at the start of PhD, I explored different labs and projects
with my early rotations establishing other pre-existing methods on Terra for
scalable
analysis of cancer samples, one leading to a manuscript. This was great for learning how to perform scalable
bioinformatics analysis, though I wanted to further learn how to develop these statistical
methods, which I then did to develop a method for copy-number variant calling from
single-cell DNA-sequencing. While methods development got me closer to the
science, I still
felt disconnected from the science due to being unfamiliar with the data generation and
experimental design, which
is a common weakness in bioinformaticians. Therefore, I realized I needed to be in the wet
lab learning how to design experiments and how to generate the data.
Now transitioning into my primary PhD project, it represents my proudest project as
I
worked on
the project from start to beginning – experimentally establishing the
cell-lines, genome editors, high-throughput screening approach, experimental design,
etc., computationally establishing the analytical framework, and
scientifically leading next-steps on exploring biological results or therapeutic
opportunities.. In fact, by having knowledge in both the experimentation and
computation, this leads to better informed experimental design (due to predicting the
computational/statistical analysis and understanding what leads to higher statistical power)
and better computational analysis (due to better
understanding of the underlying data and possible confounders/noise). Additionally, by being
directly involved in the entire scope of the project, this leads to being ruthlessly
"results-oriented", i.e. performing the experiments necessary to answer the most interesting
scientific questions and performing the analysis required to answer the scientific question,
in contrast to doing unnecessary/ill-designed experiments or doing unnecessary computational
analysis (or analysis that provides minor benefit to answering any scientific question).
Therefore, this
project represents my ultimate scientific
demonstration, with
full understanding across the entire scope leveraging my entire background
in biology (for
biological interpretation and experimentation), computer science and mathematics (for
methods development), and software engineering (for tool development) to identify novel
regulatory mechanisms of fetal hemoglobin induction and therapeutic genome editing
strategies for
sickle-cell disease and beta-thalassemia. Unfortunately this project is not
published yet and contains confidential results regarding the therapeutics, therefore only
brief details of the project will be given.
Project Description
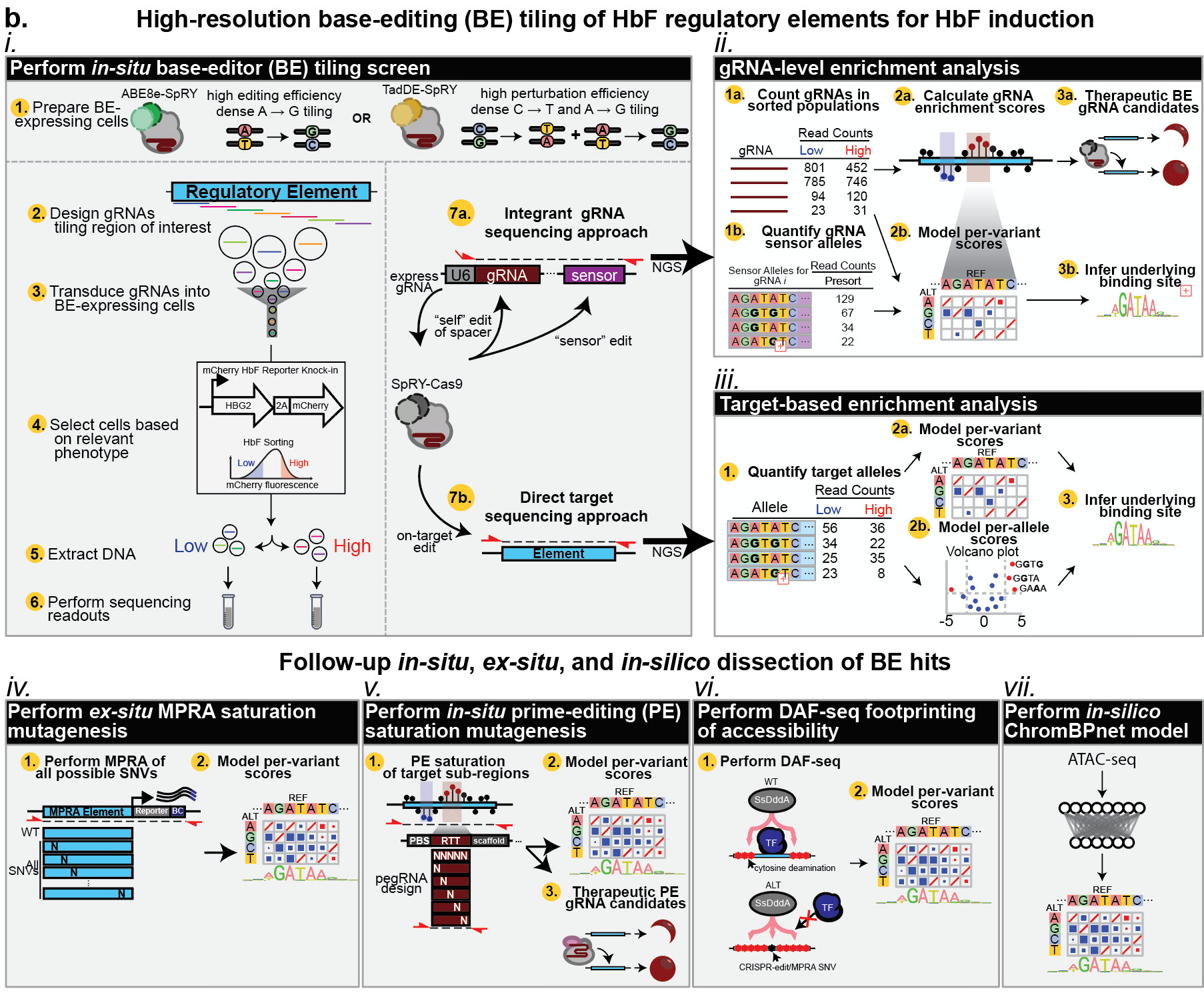
Schematic of multi-modal framework for dissecting the fetal hemoglobin regulatory sequences.
Transcriptional regulation by non-coding elements and their associated DNA-binding transcription factors (TFs) control the developmental transition from fetal to adult hemoglobin – commonly referred to as “hemoglobin switching”. The reactivation of fetal hemoglobin (HbF) has emerged as a therapeutic avenue for β-hemoglobinopathies such as sickle cell disease and β-thalassemia. Delineating the cis-regulatory logic of sequences governing HBG1 and HBG2, the genes encoding the γ-globin chains of HbF, is essential for understanding hemoglobin switching and developing more effective treatments. While population genetics studies have pinpointed single-nucleotide variants (SNVs) linked to HbF induction, this approach is limited to naturally occurring variation, preventing a comprehensive view of causal regulatory elements. To overcome this limitation, we have performed a systematic comparison of multiple mutagenesis approaches on HbF regulatory sequences in immortalized erythroid progenitors (HUDEP-2) to elucidate the variant-level cis-regulatory logic and corresponding TF binding sites underpinning HbF induction. Specifically, we integrated four complementary approaches: in-situ dense adenine- (ABE8e-SpRY) and dual- (TadDE-SpRY) base-editing mutagenesis, in-situ prime-editing saturation mutagenesis, ex-situ massively parallel reporter assay (MPRA) saturation mutagenesis, and in-silico ChromBPNet saturation mutagenesis. These approaches were performed on the HBG1/2 promoters, BCL11A +55 kb enhancer, and BCL11A +58 kb enhancer given their therapeutic relevance for potent HbF reinduction. This multi-modal screening strategy identified several novel insights into hemoglobin switching and identified multiple novel therapeutic editing strategies. This comprehensive functional mapping obtained across multiple modalities establishes an example of the strengths, weaknesses, and opportunities to combine in situ, ex situ, and in silico approaches to determine the function of regulatory elements as well as provide an optimized therapeutic roadmap for gene therapies.
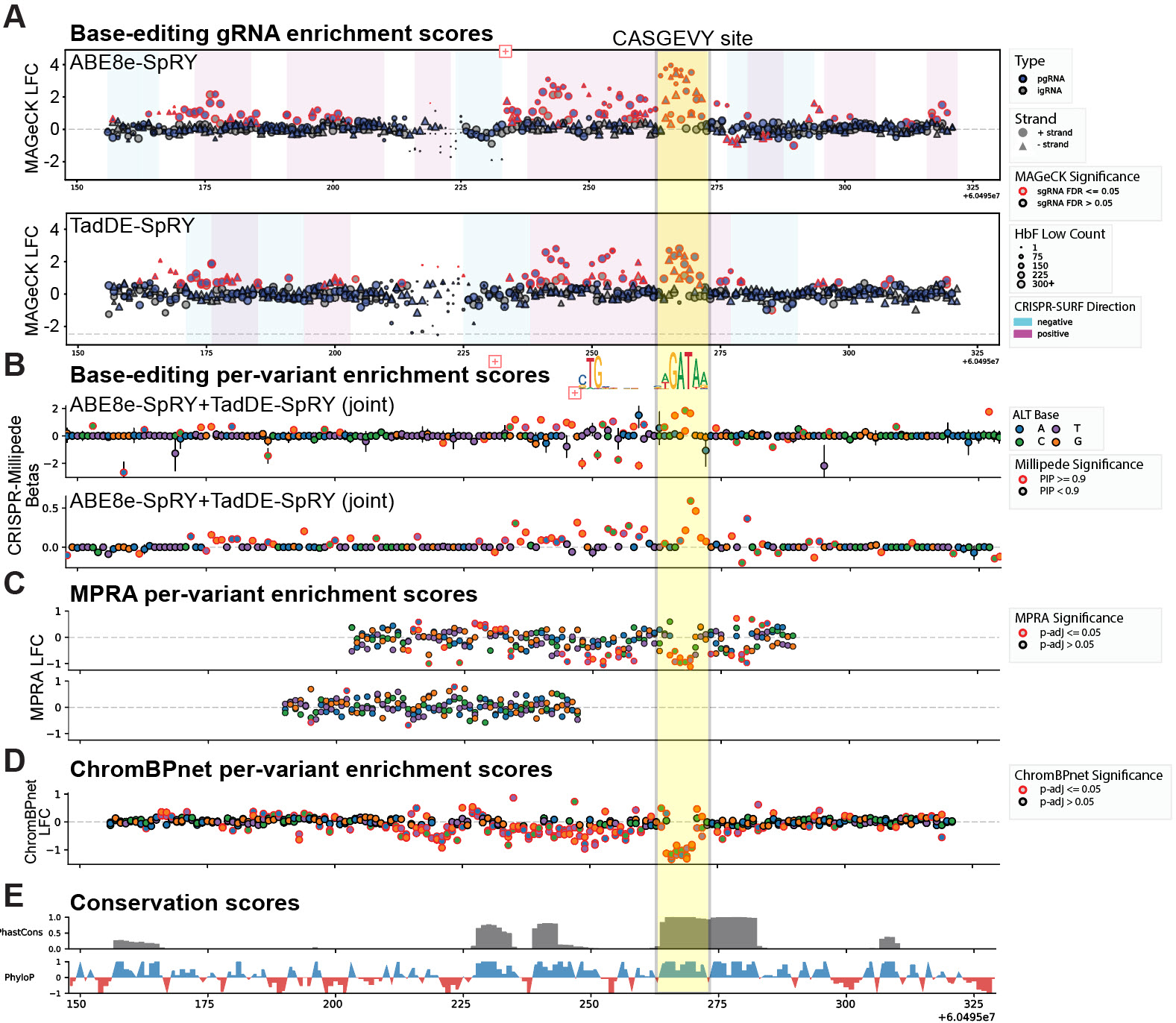
Example of multi-modality results on BCL11A +58 enhancer. Not showing BCL11A +55 and HBG2 Promoter results due to confidentiality.